Fixation
- Wash cells by centrifugation (e.g. 200 x g, 5 min, 4°C) in protein-free buffer, such as Phosphate Buffered Saline without Ca+2 or Mg+2 (PBS).
- (Optional) Repeat step 1.
- Resuspend at 2 x 106 cells in 1 ml ICE COLD BUFFER. Cell number will effect staining quality!Optional: Use pre-coated or silanized polypropylene tubes to minimize sticking. Pre-coat tubes overnight with 2% Bovine Serum Albumin (BSA) in PBS.
- Vortex gently, slowly adding the cell suspension dropwise to 9 ml of 70% ethanol in a 15 ml polypropylene centrifuge tube (Falcon® Cat. No. [35]2097). OR: Vortex gently, slowly adding the cell suspension dropwise to an equal volume of COLD ABSOLUTE ethanol. Optional: Observe cell preparation with a microcope to verify minimum cell clumping.
- Store at 4°C to - 40°C for AT LEAST 2 hours, 12 - 24 hours is best. Can be stored for up to 2 years before staining.
- Centrifuge cells at 200 x g, 10 min, 4°C.
- Resuspend pellet in 3 ml COLD PBS and transfer to Falcon® 12 X 75 mm (Cat. No. [35]2054) polystyrene tubes for staining if other tubes (polypropylene) were used for the fixation steps above. Falcon® Cat. No. [35]2235 have nylon filter caps and will remove clumps.
Staining with Propidium Iodide (PI)
- Wash cells at least once with COLD PBS. Cells may form a diffuse ring-shaped pellet, so centrifuge longer ( e.g. 200 x g, 10 min, 4°C).
- Resuspend cells in 300 - 500 µl PI/Triton X-100 staining solution: to 10 ml of 0. 1 % (v/v) Triton X-100 (Sigma) in PBS add 2 mg DNAse-free RNAse A (Sigma) and 0.40 ml of 500 µg/ml PI (e.g., Roche). Prepare freshly. A stock solution of PI, made by dissolving 1 mg PI in 2 ml water, can be stored several months at 0° to 4°C. (Or buy 500 µg/ml PI from Roche new Catalog # 11348639001, old Cat. No. 1348639). Note: If the RNAse is not DNAse-free, boil a solution of 2 mg RNAse A in 1 ml water for 5 min. Aliquot and store at -20°C.
- Incubate 37°C for 15 minutes or for 30 min at 20°C.
- Transfer tubes to ice or store at 4°C PROTECTED FROM LIGHT.
- Acquire data on flow cytometer within 48 hours (but might last up to 2 weeks). May require nylon mesh filtration (eg, Filcons, BD Cat. No. 340627) to remove cell clumps or syringing (25 gauge, UCSD Storehouse # 7245) to break up cell clumps. Can acquire 5-30 samples per hour, depending on cell preparation.
- MulticycleAV (IBM-PC) or ModFit LT (Macintosh) is used to fit the data to various cell cycle models. See below for examples
This a screen shot of a typical profile from MulticycleAV:
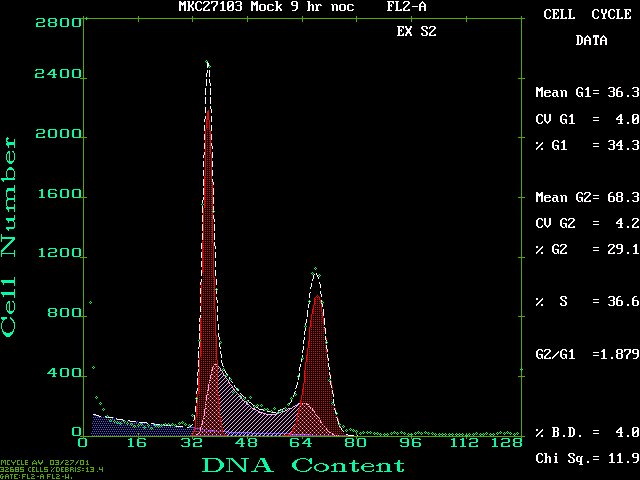
Click on this figure to see the full screen.
Here's a ModFit LT output example from the same FCS file:
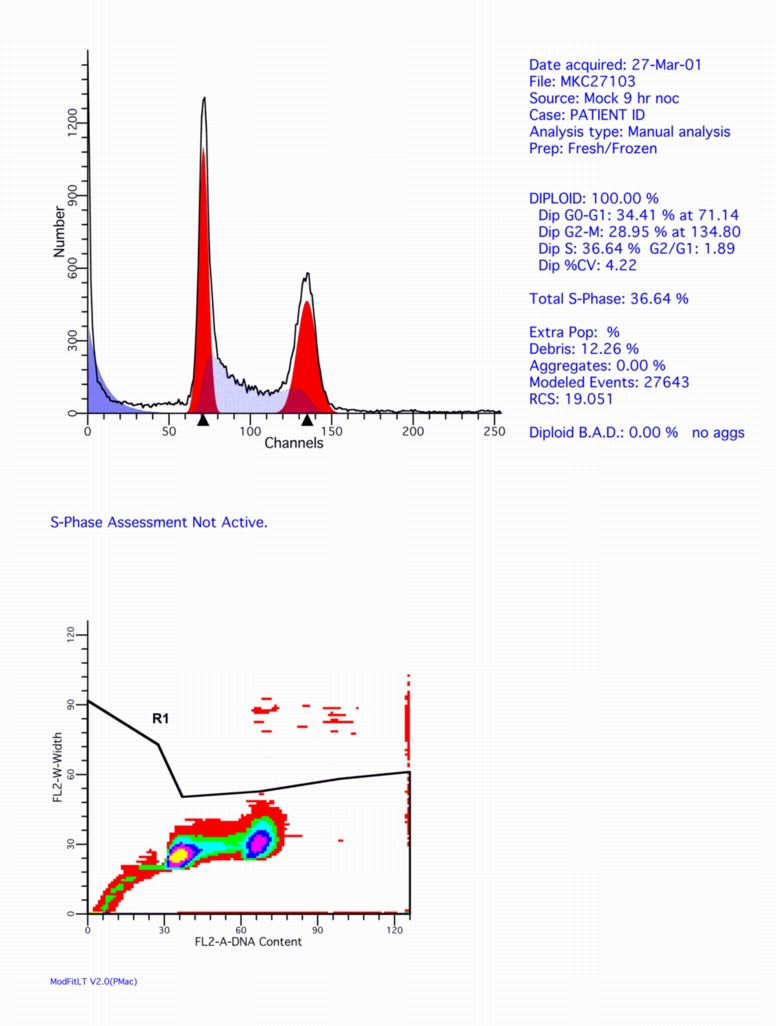
Click on this figure to see the full screen.
References
- Shapiro, HM, Practical Flow Cytometry, second edition. New York: Alan R. Liss, Inc; 1988. 353 p.
- Darzynkiewicz, Z, Nucleic Acid Analysis. In: Robinson, JP, managing editor. Current Protocols in Cytometry. New York : J Wiley & Sons, Inc; 1997. Chapter 7.